Hi there again! Week 5 was a busy one, which is why I’m writing this post on Thursday of Week 6. But don’t worry, a lot will happen between now and Sunday, so my Week 6 post will be just as action-packed as this one is. Since my internship is coming to an end, I’ve been working long hours to finish my simulations and data analysis. I wrote my very first scientific abstract (besides the ones from science fair), and now I am preparing a conference poster (much more awesome than a science fair board). Next week is going to be super busy, as I have a powerpoint presentation to give to a big group of scientists and also a poster session to attend to show my research to others. I’ve been doing a lot of practice presentations and I did a speaking competition, so my public speaking skills have improved tremendously, and I’m beyond excited for next week. My abstract will be handed out to all the scientists who attend my presentation, so I had to make sure it was perfect before I submitted it!
This pic shows how polyalanine behaves in different solvents:
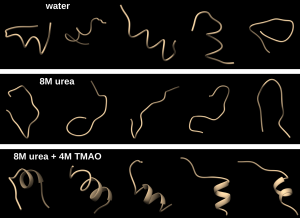
Here’s a summary of my research: I ran molecular dynamics computer simulations, using a program called GROMACS, of the amino acid alanine, in monomer and polymer form, in different solvents: pure water, urea-water, and urea-TMAO-water. This took up a huge chunk of time because simulations usually take 3 to 4 days to run using 24 computer processors. Once the simulations were finished, I would calculate their radial distribution function, which shows how much an element wants to stick to another element in a given solvent, for example how much alanine wants to interact with urea in a cosolvent of urea-water. For pentadecalanine (polyalanine with 15 alanine monomers), I calculated the radius of gyration and the end-to-end distance of polyalanine, which shows how elongated or folded the polymer is. Finally, I performed cluster analysis, which groups the different structures of polyalanine into groups, to provide a general model of what polyalanine looks like in solvents. By understanding how urea and TMAO affect proteins, we can relate this to how other substances affect proteins in general. Also, many diseases are linked to abnormal protein folding, so understanding protein interaction with the environment will help us have a deeper understanding of protein-related diseases.
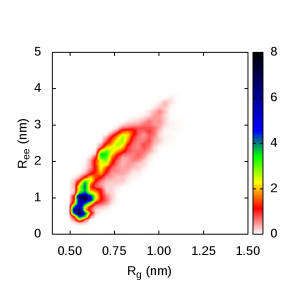
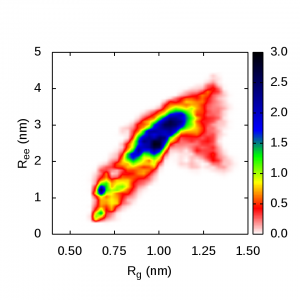
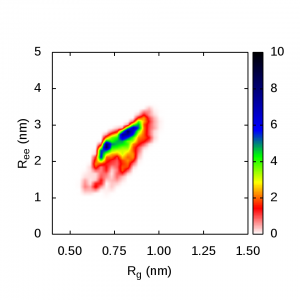
These are plots of the end-to-end distance versus radius of gyration for polyalanine. The blue regions in these pics show the highest probability of polyalanine structure in different solvents:
Polyalanine in water is compact and helical:
Polyalanine in urea is elongated into an extended or hairpin structure:
Polyalanine in urea-TMAO assumes it’s folded and helical shape again:
Basically what we were trying to find out in this research was the effects of urea and TMAO on the amino acid alanine. We chose alanine because it is one of the simplest amino acids to study. Through the simulations, I found that urea elongates polyalanine and TMAO counteracts urea’s effect by folding the polymer back into its helical form.
Here’s my little brother and sister at the Mission:

My family visited me this weekend, which was really fun. I took them to all my favorite spots and restaurants, like The Habit (the #1 burger restaurant in America!!) and McConnell’s ice cream (the absolute best ice cream I’ve ever tasted!!). We went to the beach, biked, went to Mass at the Mission, and did a lot of other fun stuff. I also got to show them around UCSB and IV, and show them my simulations, which was exciting! Oh and also this week was Fiesta, which is Santa Barbara’s biggest festival of the year. It is a Spanish festival, and there are a countless number of events such as Spanish Dancing going on from Wednesday to Sunday. My host family took me to a pancake breakfast which was marvelous! I highly recommend Fiesta because I had a blast! There are so many fun events to enjoy!!
So to conclude, I just want to say how great this research experience has been. When I first arrived in Santa Barbara, I had no idea what I was going to be researching or how much fun I would have. I had never before performed a computer simulation, and now I can successfully navigate through a LINUX terminal and perform simulations completely on my own. I have a much deeper understanding of the competition between urea and TMAO, and also protein folding in general. My mentor, Pritam Ganguly, and faculty supervisor, Joan-Emma Shea, have been amazing and I’ll be forever thankful for the knowledge they have given me. Through all the presentations and speaking competitions I’ve done at UCSB, my public speaking skills have improved tremendously. I’ve made great friends, had a blast, and changed as a person. This internship has been great practice for college, both through the social nature of it and also the research side, because I definitely want to continue doing research in college. It’s seriously so depressing that I only have two more blog posts.



There are no comments published yet.