I am Aubrielle Weaver, spending my summer in San Diego, California, at San Diego State University (SDSU). For six weeks I have been studying microbiology. My mentor is Gregory Burkeen, who is a Ph.D. student at SDSU. This is week 5 of my internship. I encourage you to read my previous blogs in order to understand more of what I’m doing.
This week was a lot of repeating previous tests and tasks. My two goals where to find a phage through a plaque assay and to get a high concentration of DNA from a DNA extraction. I needed the high concentration because another student in the lab was doing a test to see the genome of the bacteria. He was doing the test on Wednesday so I had a deadline to get good extraction. This week for each day I made a list of the things I wanted to do. This helped me keep track of everything going on with each of my bacterias and helped me write this blog. Normally I write parts of it when I have downtime at the lab to keep everything fresh. But I did not have much downtime this week. I will also be using the names I have given the Bacteria so it does not get confusing. The bacteria names are CK1, GK3, GK4. My GK1 is the fuzz that I freezed last week. I don’t need to work with it anymore. So it won’t be talked about much. My GK2 died so I haven’t been able to work with it. CK1 came from cows milk kefir. GK3 and GK4 came from goat’s milk kefir.
On Monday I started off with making a subculture for my GK4. I wanted to make the plaque assay that got pushed to Monday because I forgot to make a subculture on Friday. I already had the enrichments ready. The enrichments were from my frozen yogurt sample and the blueberry kefir sample. Named FY and BK respectfully. I had over the weekend left out the rest of my frozen yogurt sample with some kefir added into it. Ant had gotten into this sample which is a good thing because they introduced more bacteria and potential phages into the sample. It also kept them from getting inside our pantry. I didn’t end up processing the sample on Monday though. I did however make media because Greg had an autoclave time at 10:30. So I made 300mls of media and put it into the autoclave. While I waited for that I filtered my GK4 enrichment to be ready for my plaque assay. When it was finally time to check on my subculture at 2:00 pm it wasn’t ready. So I waited for longer until 4:30 and it still was not ready. So I decided to try again the next day.
The next day I made a subculture for Gk3 and CK1. Greg suspected that GK4 liked growing with no oxygen. So Greg found me a candle jar to use. He had explained what it does earlier to me when he was using it for his own work. A candle jar works like this. I took my GK4 liquid culture and put the tube in a beaker within the candle jar. Greg then secured a little candle within the jar. He lit the candle and put the lid on top which seals it tight. Then he screwed on the handle to secure the lid and provide a way of carrying it. The flame slowly consumes all of the oxygen and releases carbon dioxide or carbon monoxide. Once the flame goes out you know there is no more oxygen left. It is super satisfying to watch the flame go out slowly.
We took the candle jar along with my other two subcultures to the incubator. I then centrifuged the frozen yogurt kefir sample. I named this FYK. Once I got just the supernatant of my sample I continued with a DNA extraction. The DNA extraction got very confusing for me. I am working with likely gram positive bacteria. Which means their cell wall is thicker which results in it being harder to lyse. Lysing is breaking apart the cell to access the DNA. But since I was working with difficult bacteria we tried so many different ways with bad results. I don’t remember the order of what we tried but I will explain each method. The first way is freeze thaw. So we took an ice bucket and grabbed some dry ice. We placed my bacteria sample onto the ice until frozen, then we placed it into this heating machine until it was thawed. And repeated that multiple times. We were looking for the sample to clear and become viscous. Which means it is more like syrup than water when pouring.
Another way is adding detergent like SDS to dissolve the cell membrane and protease K to disrupt the cell wall. You incubate this on a shaker at 37 degrees celsius. Another way is bead beating. This is when you add little beads into the sample and place it onto a shaker, or a thing that turns like a ferris wheel. The beads create bubbles which disrupts the cell wall. None of these ways truly gave me lysis but I still attempted to get DNA from centrifuging and filtering. None of this worked so overnight we left it bead beating to see if that would help. Once my subcultures were done I added phage samples to my CK1 and GK3 for enrichments.
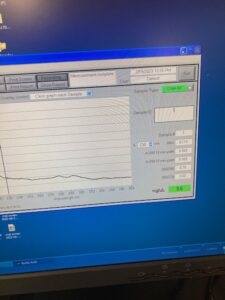
The next day I observed that my GK4 samples grew a lot more than they typically do. This was great news. So I made a subculture for all three bacterias. I wanted to do a plaque assay for CK1 and GK3 and make an enrichment for GK4. So I got my subcultures going and continued with the DNA extraction. Once we got some lysis we took it to a machine called a nanodrop. This measures the concentration. You put two micro liters of water onto the little dot and run it to get a control measure. Then you put two microliters of your sample on and run it. It tells you how many nanograms per microliter you get. The highest I ever got is 18.5 ng/ul and that isn’t very high. The machine is also not very accurate. Once my subcultures were done I made my plaque assays and went home.
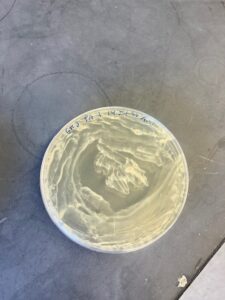
On Thursday I didn’t have any plaques. So I just made my subcultures for each bacteria, processed my GK4 enrichment and made plaque assay plates for it. And made a GK3 FYK enrichment. My CK1 plate had a potential plaque but likely not. I still tested it though and streaked it onto another plate.
The next day revealed that I didn’t have a plaque. I had made a subculture and was streaking my bacteria forward while I waited for Greg to come back to take me to the incubator, when the professor of the lab, Forrest Rohwer, came back from his trip and called a lab meeting. I didn’t know he was coming back so it was a bit of a surprise. He asked Rachel and I to talk about what we were doing. Rohwer wanted me to make a hypothesis even though I can’t test it and I will talk about that to the group again tomorrow. Once the meeting was done I took my subculture to the incubator and made liquid cultures for each of my bacteria. I also made a little bit more plates and sterilized some DI water.
Thursday and Friday night we went to watch the sunset on the beach for Natalie’s and Mel’s last days. I also went to the beach to swim and read for a bit on Saturday. Later today I am going to see the new Barbie movie. I look forward to one more week!




There are no comments published yet.