I am Aubrielle Weaver, and I am spending my summer in San Diego, California, at San Diego State University (SDSU). For six weeks, I have been studying microbiology. My mentor is Gregory Burkeen, who is a Ph.D. student at SDSU. This is week 3 of my internship. I encourage you to read my previous blogs to understand more about my work.
Since the 4th of July was on Tuesday, I didn’t need to go to work until Wednesday. This gave the milk sample I had set outside plenty of time to rott. I hoped I could collect phages from it. The four-day weekend also allowed my bacteria back at the lab to grow.
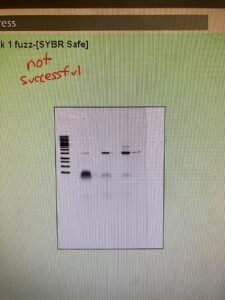
I first ran a PCR on my mysterious fuzz to confirm it wasn’t bacteria. I needed to run it through the thermocycler like I ran my other bacteria to prove it. If it’s not bacteria, no DNA should be replicated during PCR, so there won’t be a result after the gel electrophoresis. I also needed positive and negative control. The positive control contained bacteria from one of my plates. And the negative control didn’t have any samples, just the PCR ingredients. However, my results showed up with each row being positive. That means I had contamination. This was frustrating because it took over two and a half hours to get results. I had to redo the PCR completely fresh. I sanitized the micropipettes, got new micropipette tips, used a new Taq Polymerase and thermocycler water, and made a new concentration of primers. I made a new positive and negative control, along with the sample I wanted to test. I left them in the thermocycler overnight.
While waiting for my first PCR, I made subcultures from three liquid cultures and prepared my rotten milk sample. The fourth liquid culture had no growth, so I created a new one. I did a one-to-fifty subculture to have enough growth sooner. I had to wait for a couple of hours for them to grow. I then centrifuged my milk sample and moved it into new tubes until I had no chunks in the sample. Once the subcultures were done, I made my enrichments. I split my milk sample equally amongst the subcultures, leaving enough for the other liquid culture that I needed to redo. And they were sent to the incubator.
On Thursday, I came in and immediately made my gel for the redo test on my fuzz. I had to use a different buffer because I ran out of the one I had been using, and Greg hasn’t shown me how to make it yet. He will later on. And then, I retrieved my subcultures from the incubator. I then streaked one of my bacteria to keep it alive and then put some glass tubes into the autoclave for ten minutes to sterilize. Once sterilized, I made subcultures for my three bacteria and a liquid culture for my fourth bacteria. Those were sent to the incubator, and I then checked on my gel.
The results from my gel were the same as the first time. That means either the primers are contaminated, or I need to improve at aseptic technique. Greg said it’s likely that the primers are contaminated since that is the one thing we didn’t change. Yesterday I made a new dilution of primers. Still, I used the same primer shipment as the original primer dilution I was using. So with new primers from a different lab, I redid the PCR for the third time. I then took my subcultures from the incubator and started my plaque assays. This is where I have a control plate to confirm my bacteria is growing evenly and a test plate with bacteria and my phage samples. I have a phage if I get circular zones (plaques) with no growth. I did this test last week but was unsuccessful. Like last week, I had control and test plates. But I needed two plates per bacteria since I worked with three different bacteria and only one phage sample (rotten milk). Both control and test plates had the subculture bacteria spread on. Then once they were dry, I took my enrichments and centrifuged them at full speed for a minute. I took 1 milliliter of the supernatant and moved it into the filter tube, carefully not picking up the bacteria pellet. I then centrifuged those at full speed for a minute. Once filtered, I dumped the filter sample onto their respective test plates. Once dry, I placed it into the incubator.
On my way to SDSU the following day, I realized I had forgotten to put samples into my PCR tubes. I informed Greg, and he said it happens more often than you think, so don’t worry about it. I first grabbed my plates and liquid culture from the incubator. The liquid culture had no growth. This likely means that the bacteria have died. I streaked it from one plate to another to try and get more growth. My plaque assay plates had no plaques. Greg explained that I likely have phages; they are just not the type that creates zones of clearing. So I might do a newer test to find phages. After observing my plates, I created my new PCR tubes. I made sure to put samples in this time. While I was waiting for my PCR, Greg noticed a fruity smell coming from two of my plaque assay plates—the control and test for one of my bacteria. The original plate of this bacteria didn’t have this smell. Since only one of the plates has the phage sample, the smell is not different because of the phages. I streaked forward the fruity bacteria to see if the scent would stay.
Once my gel electrophoresis was done, we looked at my results. My positive control and negative came out right, and bacteria are in my fuzz. Because of the potential contamination, I had in my earlier PCRs, I need to redo the other four bacterias I had run. So I prepared those and started the PCR. I can leave those over the weekend because the last temperature the pcr stays at is fridge temp. So they can sit in there for a while.




Aubrielle, this is all so interesting, and it sounds challenging as well! If you were to repeat this testing on the rotten milk, would you leave it out for longer or do anything differently? What are you doing with your free time outside of your internship? Are you getting to explore the area and see new paces? I can’t wait for next week’s update!
Greg has mentioned a potential new test I might do to find phages in my rotten milk samples. This past weekend I left out more dairy products to try and find phages. I didn’t do much for free time. I just chilled around the house, worked on a paint by number kit, and I watched the sunset after I had posted my blog. It was a calmer week. The past few weeks I’ve explored the La Jolla area more but haven’t gone to down town San Diego yet.