Hey everybody! Welcome to my third blog post! This week my internship switched focus to another one of the many interesting techniques used in Computer Aided Drug Discovery. Instead of continuing to learn the ins and outs of Rosetta with Vladimir Yarov-Yarovoy-Yarovoy, we are learning about molecular dynamics with our second mentor, Igor Vorobyov. We started out the week with a zoom call introduction where I was introduced to Slava, a colleague of Igor, and his two student teachers, John and Kyle. To finish the first meeting Igor gave us a lecture he gives to his students. It introduces molecular dynamics, which I have come to define as a computational technique involving the evolution of time. This brought our introduction to Computer Aided Drug Discovery to a whole new level. Now, not only were we introduced to a technique that could show how a drug interacts with a protein at a specific moment, we also are able to see their relationship over time. The next day, our meeting focused on installing the software used for molecular dynamics. We were all having some tech issues so we weren’t able to get everything sorted out. The next morning I met with Kyle to figure out why the software wasn’t working properly. It turned out that the new MacOS update wasn’t compatible with what we were doing so we had to download a later version of the software.
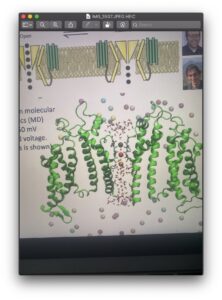
At our next meeting we took a break from what we were doing and listened to another one of Igor’s lectures. This one focused on Cardiovascular System Modulation. The goal of this research, done by Igor and many of his colleagues, was to make prescription drugs that cause heart arrhythmias more safe. Around 80% of drug discoveries are shut down before they make it to clinical trails because they have the potential to cause an arrhythmia know as torsades de pointes. The drugs are tested for a hERG channel inhibition and automatically shut down if one is present. The hERG channel is responsible for moving ions across cardiac cell membrane. The issue is not all hERG inhibitors are pro-arrhythmic. So, Igor and his team tried to solve this issue by computational how the drug moves through the membrane. Though I only got a brief idea of the things going on in Igor’s lab, I am so glad to have the ability to learn about what C.A.D.D is being used for in the field. That was all we got around to doing this week as we had some various technical issues. Below I will attach an image of the hERG channel that was modeled computationally. Thanks for reading and see you next week!

There are no comments published yet.