Welcome to Week 5 of the Computational Biology Internship. My name is Eliram Reyes-Powell and I am interning at University of California, Davis, School of Medicine in the Department of Physiology and Membrane Biology exploring Molecular Dynamics with Dr. Igor Vorobyov, Dr. Slava Bekker, and PhD Candidate John Dawson as the second section of the internship.
This is the last week of two very challenging and fruitful weeks. Next week, we will move on to Dr. Colleen Clancy’s Laboratory to complete this internship experience (Which has involved a progression in which the findings of one field are applied to the other, something very interesting and enlightening as to what the world of science really looks like).
This week started with background information about Cardiac Arrhythmia, a very concerning and common ailment in the United States. A very surprising statistic was that about 50%-70% of all small molecule leads (drugs candidates) are discarded because they are pro-arrhythmic (they cause arrhythmia). That was something I did not expect to hear, and it makes me very concerned that, perhaps, many of the drugs that we consume have similar effects, but they do it at a slower rate (do not be alarmed, it was just my first impression). For that type of problem or question is when Molecular Dynamics simulations and analyses can take place and one of the reasons many UC Davis laboratories (and others around the world) are collaborating to investigate them in-depth.
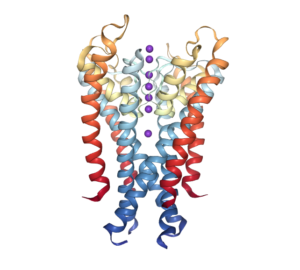
On that same day, we went over LQTS (prolonged action potentials), the polarization of membranes and how that causes channels to open or close, different ways in which ions can be transported (not only the ion channels I have been exploring, but ion transporters), how selectivity filters behave, and a more detailed introduction to analysis techniques. I was also introduced to the alpha subunit (Kv11.1) of the potassium ion channel which is coded by the gene hERG (which is the name used to refer to the protein section). All of that information gave me a broader sight of the scope for advancement available in the field, as well as a more educated perspective on all of the information I receive from my mentors.
Visualization of hERG (Kv11.1):
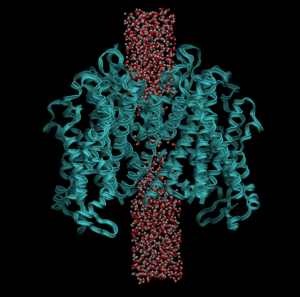
On Tuesday, we explored further the selection language used by VMD. Mr. Dawson explained how he visualizes ion channels and the “sentences” used for the program to select specific regions of the protein and its environment. I followed along with his actions and I got excellent results from the simulation and analysis. It was a 300 nanosecond simulation with many events affecting the system at the same time, so specificity was very important. Mr. Dawson, then, showed some of the coding (extremely advanced, to say the least) that is used to build the plots using the millions of data fragments from simulation into a common graph (used for comparison). I was very impressed and amazed by the results, so I want to learn how to build such colorful and complex graphs as soon as possible.
Visualization of the protein channel with a cylindrical selection of water molecules:
On Wednesday, we, the interns, did a very fun activity. We were asked to count the number of potassium ions that go through the selectivity filter of the Open hERG during a 1500 nanosecond simulation and to obtain the conductance value to see if the simulation was appropriate. We spent some time debating about what counting method to use and which result was the most accurate. Our final answer was eight potassium ions. From that number, we used Ohm’s Law (having the Voltage, given, and Current, calculated by us, variables available) to obtain Resistance and use that value to obtain the Conductance of the ion-channel.
After discussing the results with our mentors, the conclusion was that the selectivity filter was very selective for potassium ions, the selectivity filter wasn’t aligning correctly (may have had an altered sequencing), and there was not enough water flow to allow more potassium ions to travel through the channel effectively. It was great to hear my mentors’ feedback and their conclusions about what my and their results indicated about the system. Before wrapping up for the day, Mr. Dawson commented that in vitro (involving isolation of real molecules, cells, etc.) experimental evidence would be the next step to prove the in silico (computer based) results. Of course, I can’t work with in vitro protocols due to the current situation, but I can learn about the process which is very exciting.
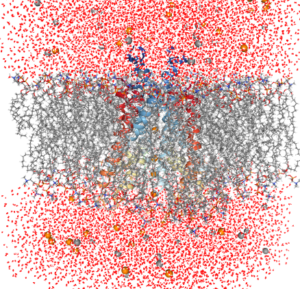
On Thursday, Dr. Slava Bekker and Dr. Igor Vorobyov introduced the CHARMM-GUI, Membrane Builder, which is used to build a protein/membrane complex for molecular dynamics simulations. For the hERG simulations we did on the previous days, the professors had given us membrane-included versions, generated with CHARMM-GUI. However, they wanted us to know how they obtained the membrane and water molecules so we can do it in the future. It is quite complicated to do and requires experience, so I am glad I was able to learn how. We wrapped up the session by quickly performing all of the protocols we had been working on throughout the days in a few minutes. It was great to see how proficient I have become with using VMD. I still have a long way to go, but the improvement is evident.
Here is how hERG looks when having a membrane and water molecules generated around it with CHARMM-GUI:
I have been able to learn a lot about the physics that govern the molecular world. It has certainly been challenging, but my mentors have been very committed and all of the knowledge that they share with me makes me realize how amazing molecular dynamics is and the long road I still need to travel in order to be proficient with all the concepts, techniques, and analyses they do.
I am very grateful for the opportunity and I look forward to learning more about Computational Biology (Molecular Modelling and Molecular Dynamics so far). Thanks to all of my mentors: Dr. Vladimir Yarov-Yarovoy, Mr. Brandon Harris, Dr. Igor Vorobyov, Dr. Slava Bekker, and Mr. John Dawson for all of their support, kindness, and great experiences.
When not in my internship, I spent some time studying for the SAT, advancing in my AP Calculus BC course (I need to finish the first semester of it before school starts), and (although I haven’t finished the book “Six Easy Pieces”, by Richard Feynman) I started to go over a Linear Algebra book my grandfather gave to me, so I am very curious about that branch of mathematics now.

There are no comments published yet.