I hope I find you all safe and healthy!! My name is Eliram Reyes-Powell and I am doing a virtual internship, through Pinhead Institute, at the University of California, Davis School of Medicine in the Department of Physiology and Membrane Biology. This will be a two-part internship, with the first part being in the Protein Modeling field with Dr. Vladimir Yarov-Yarovoy and PhD Candidate Brandon Harris.
For this internship, I have the good fortune to collaborate with another 2020 Pintern, Kelly Stellmacher, as well as with other two students. We have been supporting each other throughout the preparation weeks as well as along this first week of the internship. Since collaboration is a very important skill in science, it is a great experience to be working with other students on the same projects.
The biology knowledge and computational skills needed for this internship are well beyond high-school level, so I have had to study many hours in order to understand how proteins behave and how their structures determine their functionality. Also, using the SSH and command line interpreter to run Rosetta (for protein modeling) have been very enriching experiences and a great introduction to computational biology.
To have a better visualization of protein interactions, we started to play a video game called Foldit, which Dr. Vladimir Yarov-Yarovoy recommended we try. Through this game I have learnt about how proteins relax, about the secondary structures of proteins, and many processes proteins undergo. However, the most interesting feature of this game is that by playing it, you can contribute to the discovery of new proteins and their lowest energy state, thereby helping find solutions to many problems in the world of medicine. So, I intend to continue to play after the Internship is done.
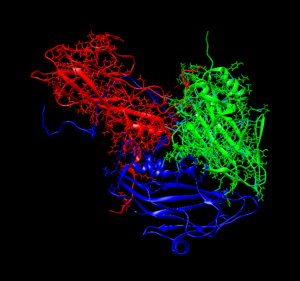
After gaining the necessary knowledge to understand proteins and their molecular interactions, I accessed the Protein Data Bank. In the Protein Data Bank (PDB), with the instruction of Mr. Brandon Harris, I learnt how to download the PDB data files of different proteins to, then, graphically visualize them using the UCSF Chimera software. After that, Mr. Harris taught me the protocols to correctly prepare proteins for their usage in Rosetta Software, which I will talk about next week.
6P9O Virus Structure Using UCSF Chimera:
When we were getting some context on the field and on what the models can be used for, I was able (through a TED Talk by Professor David Baker) to realize how developing new proteins could be a key for the FUTURE:
Humanity has discovered a minuscule fraction of all the available protein combinations (Which depend on orientation, size, and just random arrangements of amino acid sequences). The amount of proteins that can be discovered is astronomical (20^100 to be exact). So, as you might imagine, the functions of the different proteins are almost limitless.
By discovering new proteins, many pharmaceuticals can be created, the cures for lethal illnesses could be developed, extremely strong materials could be manufactured, and many aspects of modern medicine would change.
It is very encouraging to know that some of the problems we are going through right now could be solved by modeling and manufacturing protein functions. Molecular Modeling is a field that could eventually become very advantageous for humanity. So, I am very grateful for the opportunity to learn about the field and contribute to it.

There are no comments published yet.