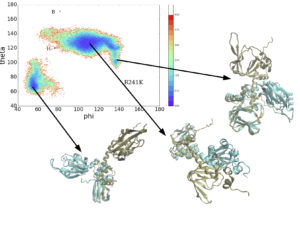
This is a graph that shows the probability of certain conformations happening. It shows what the things on the graph actually look like.
Week 3! This week has been long and slow for me, but exciting! For the past week I have been waiting for my simulations to finish processing. I have gathered about 300 nanoseconds of simulation from the supercomputer. The complexity of the process means that it often takes a while to get the simulations. A nanosecond is one billionth of a second, or .00000001 seconds. It takes an entire day to get .00000027 seconds of footage; which is actually quite fast. When I get all of my footage I will begin doing data analysis for my project.
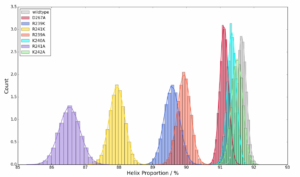
While I am waiting I have continued helping my mentor Emilia with her project. We have been doing a lot of data analysis on some mutations of protein kinase A. I ran several programs in order to create different visual representations. In total I created around 15 different graphs. I learned a lot about computer coding this week, mainly using the programs Python and Jupyter Notebook, which was how I created all of the graphs.
Outside of the lab I have been hanging out with my host family. My host mom hurt her foot so it has become my job to walk the dogs when she can’t; I enjoy getting out of the house. I also went to the movies and saw Finding Dory, which was awesome! Two other Pinterns are joining me this weekend at my host family’s house. I am excited to have some new roommates!

There are no comments published yet.